| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:39:19 UTC |
|---|
| Update Date | 2020-05-21 16:28:32 UTC |
|---|
| BMDB ID | BMDB0000972 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | N10-Formyl-THF |
|---|
| Description | 10-Formyltetrahydrofolate, also known as 10-formyl-THF or 10-formyl-H4pteglu1, belongs to the class of organic compounds known as tetrahydrofolic acids. These are heterocyclic compounds based on the 5,6,7,8-tetrahydropteroic acid skeleton conjugated with at least one L-glutamic acid unit. 10-Formyltetrahydrofolate is a strong basic compound (based on its pKa). 10-Formyltetrahydrofolate exists in all eukaryotes, ranging from yeast to humans. |
|---|
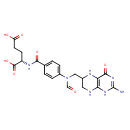
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 10-Formyl-THF | Kegg | | 10-Formyltetrahydrofolic acid | Generator | | 10-Formyl-(6Rs)-tetrahydrofolic acid | HMDB | | 10-Formyl-H4pteglu1 | HMDB | | 10-Formyl-tetrahydrofolate | HMDB | | 10-Formyltetrahydropteroylglutamate | HMDB | | 10-Formyltetrahydropteroylglutamic acid | HMDB | | 10-FTHF | HMDB | | N-[p-[N-[(2-Amino-5,6,7,8-tetrahydro-4-hydroxy-6-pteridinyl)methyl]formamido]benzoyl]-glutamate | HMDB | | N-[p-[N-[(2-Amino-5,6,7,8-tetrahydro-4-hydroxy-6-pteridinyl)methyl]formamido]benzoyl]-glutamic acid | HMDB | | N-[p-[N-[(2-Amino-5,6,7,8-tetrahydro-4-hydroxy-6-pteridinyl)methyl]formamido]benzoyl]-L-glutamate | HMDB | | N-[p-[N-[(2-Amino-5,6,7,8-tetrahydro-4-hydroxy-6-pteridinyl)methyl]formamido]benzoyl]-L-glutamic acid | HMDB | | N10-Formyl-5,6,7,8-tetrahydrofolate | HMDB | | N10-Formyl-5,6,7,8-tetrahydrofolic acid | HMDB | | N10-Formyl-H4F | HMDB | | N10-Formyl-THF | HMDB | | N10-Formyltetrahydrofolate | HMDB | | N10-Formyltetrahydrofolic acid | HMDB | | N10-Formyltetrahydropteroylglutamate | HMDB |
|
|---|
| Chemical Formula | C20H23N7O7 |
|---|
| Average Molecular Weight | 473.4393 |
|---|
| Monoisotopic Molecular Weight | 473.165896125 |
|---|
| IUPAC Name | (2S)-2-[(4-{N-[(4-hydroxy-2-imino-1,2,5,6,7,8-hexahydropteridin-6-yl)methyl]formamido}phenyl)formamido]pentanedioic acid |
|---|
| Traditional Name | (2S)-2-[(4-{N-[(4-hydroxy-2-imino-5,6,7,8-tetrahydro-1H-pteridin-6-yl)methyl]formamido}phenyl)formamido]pentanedioic acid |
|---|
| CAS Registry Number | 2800-34-2 |
|---|
| SMILES | NC1=NC(=O)C2=C(NCC(CN(C=O)C3=CC=C(C=C3)C(=O)N[C@@H](CCC(O)=O)C(O)=O)N2)N1 |
|---|
| InChI Identifier | InChI=1S/C20H23N7O7/c21-20-25-16-15(18(32)26-20)23-11(7-22-16)8-27(9-28)12-3-1-10(2-4-12)17(31)24-13(19(33)34)5-6-14(29)30/h1-4,9,11,13,23H,5-8H2,(H,24,31)(H,29,30)(H,33,34)(H4,21,22,25,26,32)/t11?,13-/m0/s1 |
|---|
| InChI Key | AUFGTPPARQZWDO-YUZLPWPTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as tetrahydrofolic acids. These are heterocyclic compounds based on the 5,6,7,8-tetrahydropteroic acid skeleton conjugated with at least one L-glutamic acid unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Tetrahydrofolic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tetrahydrofolic acid
- Glutamic acid or derivatives
- N-acyl-alpha amino acid or derivatives
- N-acyl-alpha-amino acid
- Hippuric acid
- Hippuric acid or derivatives
- Acylaminobenzoic acid or derivatives
- Alpha-amino acid or derivatives
- Anilide
- Benzoic acid or derivatives
- Benzamide
- Benzoyl
- Pyrimidone
- Aminopyrimidine
- Secondary aliphatic/aromatic amine
- Dicarboxylic acid or derivatives
- Benzenoid
- Pyrimidine
- Monocyclic benzene moiety
- Heteroaromatic compound
- Vinylogous amide
- Tertiary carboxylic acid amide
- Amino acid or derivatives
- Amino acid
- Carboxamide group
- Secondary carboxylic acid amide
- Azacycle
- Carboxylic acid
- Carboxylic acid derivative
- Secondary amine
- Organic oxygen compound
- Organic nitrogen compound
- Carbonyl group
- Hydrocarbon derivative
- Amine
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | Not Available |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cytoplasm
- Lysosome
- Mitochondria
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004j-2733900000-d416b73a1673d1220b87 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0v4i-2913374000-7b894dbf7f83fe5cd9d7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_16) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05cr-0401900000-21104c38dc03ca4e7a0e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-003r-0935700000-2f12524764eca10dcf15 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-0910000000-03f41886e7967d0201eb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0000900000-1244cea99701ed75f41c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00bc-1123900000-cdcadc51a3f47cb93886 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9842000000-62efadef6701421ce2c5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002b-0002900000-e6e9d4109ecb3ef0bbee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-1729400000-02f5fdbf97e3ce7a3a95 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001j-0932000000-68bdb6e09b80b22288c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0000900000-19d839ffa3d1b1988450 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-0200900000-93ad732aff1ff91aa085 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0v03-4920300000-17e6f1c09b0e093800ef | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|