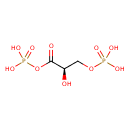| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:24 UTC |
|---|
| Update Date | 2020-05-21 16:29:05 UTC |
|---|
| BMDB ID | BMDB0001270 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Glyceric acid 1,3-biphosphate |
|---|
| Description | Glyceric acid 1,3-biphosphate, also known as 1,3-biphosphoglycerate or 3-phospho-D-glyceroyl phosphate, belongs to the class of organic compounds known as acyl monophosphates. These are organic compounds containing a monophosphate linked to an acyl group. They have the general structure R-CO-P(O)(O)OH, R=H or organyl. Glyceric acid 1,3-biphosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Glyceric acid 1,3-biphosphate exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-2-Hydroxy-3-(phosphonooxy)-1-monoanhydride with phosphoric propanoic acid | ChEBI | | 1,3-Bisphospho-D-glycerate | ChEBI | | 3-Phospho-D-glyceroyl phosphate | ChEBI | | D-Glycerate 1,3-diphosphate | Kegg | | (R)-2-Hydroxy-3-(phosphonooxy)-1-monoanhydride with phosphoric propanoate | Generator | | 1,3-Bisphospho-D-glyceric acid | Generator | | 3-Phospho-D-glyceroyl phosphoric acid | Generator | | D-Glyceric acid 1,3-diphosphoric acid | Generator | | Glycerate 1,3-biphosphate | Generator | | Glyceric acid 1,3-biphosphoric acid | Generator | | 3-Phospho-D-glyceroyl dihydrogen phosphoric acid | HMDB | | 1,3-Biphosphoglycerate | HMDB | | 1,3-Bisphosphoglycerate | HMDB | | 1,3-Bisphosphoglyceric acid | HMDB | | 1,3-Diphosphoglycerate | HMDB | | 1,3-Diphosphoglyceric acid | HMDB | | 3-Phospho-D-glyceroyl-phosphate | HMDB | | 3-Phosphoglyceroyl phosphate | HMDB | | 3-Phosphoglyceroyl-p | HMDB | | 3-Phosphoglyceroyl-phosphate | HMDB | | 3-Phosphonato-D-glyceroyl phosphate | HMDB | | 3-Phosphonatoglyceroyl phosphate | HMDB | | 3-p-Glyceroyl-p | HMDB | | D-Glycerate 1,3-biphosphate | HMDB | | D-Glycerate 1,3-bisphosphate | HMDB | | D-Glyceric acid 1,3-biphosphate | HMDB | | D-Glyceric acid 1,3-bisphosphate | HMDB | | D-Glyceric acid 1,3-diphosphate | HMDB | | DPG | HMDB | | Glycerate 1,3-bisphosphate | HMDB | | Glycerate 1,3-diphosphate | HMDB | | Glyceric acid 1,3-bisphosphate | HMDB | | Glyceric acid 1,3-diphosphate | HMDB | | Phosphoglyceroyl-p | HMDB | | p-Glyceroyl-p | HMDB | | Glyceric acid 1,3-biphosphate | HMDB |
|
|---|
| Chemical Formula | C3H8O10P2 |
|---|
| Average Molecular Weight | 266.0371 |
|---|
| Monoisotopic Molecular Weight | 265.9592695 |
|---|
| IUPAC Name | {[(2R)-2-hydroxy-3-(phosphonooxy)propanoyl]oxy}phosphonic acid |
|---|
| Traditional Name | 1,3-bisphospho-D-glycerate |
|---|
| CAS Registry Number | 1981-49-3 |
|---|
| SMILES | O[C@H](COP(O)(O)=O)C(=O)OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H8O10P2/c4-2(1-12-14(6,7)8)3(5)13-15(9,10)11/h2,4H,1H2,(H2,6,7,8)(H2,9,10,11)/t2-/m1/s1 |
|---|
| InChI Key | LJQLQCAXBUHEAZ-UWTATZPHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as acyl monophosphates. These are organic compounds containing a monophosphate linked to an acyl group. They have the general structure R-CO-P(O)(O)OH, R=H or organyl. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic phosphoric acids and derivatives |
|---|
| Sub Class | Phosphate esters |
|---|
| Direct Parent | Acyl monophosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Acyl monophosphate
- Glyceric_acid
- Monoalkyl phosphate
- Monosaccharide
- Alkyl phosphate
- Carboxylic acid salt
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid derivative
- Organic oxide
- Organooxygen compound
- Organic oxygen compound
- Alcohol
- Carbonyl group
- Hydrocarbon derivative
- Organic salt
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0002-9500000000-cb3071f9249ee92b7d2c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-1950000000-2cde7b5739871a882542 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014m-3910000000-72f2c46eab2036938056 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000f-9500000000-c835034f0ac11618922d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03fr-6190000000-6f4870543fc6131d14a7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-d9d9c6d217a255d791fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-1fef400a1ea8ca54c802 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03dj-0090000000-def40aa9bb3094302ce8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01r2-9040000000-374a9ff8c866f10f2f3c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-4bf2b9a14f4e1c8fc62a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kb-1390000000-8fe62bbfc56bf59f28a6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9410000000-e17767e21640a6f81532 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-c41dab2684b9d54afd31 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|