| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:53:28 UTC |
|---|
| Update Date | 2020-05-21 16:29:04 UTC |
|---|
| BMDB ID | BMDB0002275 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 7,8-Dihydroneopterin |
|---|
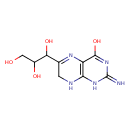
| Description | 2-Amino-6-[(1R,2S)-1,2,3-trihydroxypropyl]-7,8-dihydro-3H-pteridin-4-one, also known as 2-amino-4-hydroxy-6-(D-erythro-1',2',3'-trihydroxypropyl)-7,8-dihydropteridine or 7,8-dihydro-D-neopterin, belongs to the class of organic compounds known as biopterins and derivatives. These are coenzymes containing a 2-amino-pteridine-4-one derivative. They are mainly synthesized in several parts of the body, including the pineal gland. 2-Amino-6-[(1R,2S)-1,2,3-trihydroxypropyl]-7,8-dihydro-3H-pteridin-4-one exists in all living organisms, ranging from bacteria to humans. Based on a literature review very few articles have been published on 2-Amino-6-[(1R,2S)-1,2,3-trihydroxypropyl]-7,8-dihydro-3H-pteridin-4-one. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Amino-4-hydroxy-6-(D-erythro-1',2',3'-trihydroxypropyl)-7,8-dihydropteridine | HMDB | | 2-Amino-4-hydroxy-6-(D-erythro-1,2,3-trihydroxypropyl)-7,8-dihydropteridine | HMDB | | 2-Amino-6-[(1S,2R)-1,2,3-trihydroxypropyl]-7,8-dihydropteridin-4(1H)-one | HMDB | | 2-Amino-6-[(1S,2R)-1,2,3-trihydroxypropyl]-7,8-dihydropteridin-4(3H)-one | HMDB | | 2-Amino-7,8-dihydro-6-(1,2,3-trihydroxypropyl)-4(1H)-pteridinone | HMDB | | 7,8-Dihydro-D-erythro-neopterin | HMDB | | 7,8-Dihydro-D-neopterin | HMDB | | D-Erythro-7,8-dihydroneopterin | HMDB | | Dihydroneopterin | HMDB | | NPR | HMDB | | 7,8-Dihydro-neopterin | HMDB |
|
|---|
| Chemical Formula | C9H13N5O4 |
|---|
| Average Molecular Weight | 255.2306 |
|---|
| Monoisotopic Molecular Weight | 255.096753929 |
|---|
| IUPAC Name | 2-amino-6-(1,2,3-trihydroxypropyl)-1,4,7,8-tetrahydropteridin-4-one |
|---|
| Traditional Name | 2-amino-6-(1,2,3-trihydroxypropyl)-7,8-dihydro-1H-pteridin-4-one |
|---|
| CAS Registry Number | 1218-98-0 |
|---|
| SMILES | NC1=NC(=O)C2=C(NCC(=N2)C(O)C(O)CO)N1 |
|---|
| InChI Identifier | InChI=1S/C9H13N5O4/c10-9-13-7-5(8(18)14-9)12-3(1-11-7)6(17)4(16)2-15/h4,6,15-17H,1-2H2,(H4,10,11,13,14,18) |
|---|
| InChI Key | YQIFAMYNGGOTFB-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as biopterins and derivatives. These are coenzymes containing a 2-amino-pteridine-4-one derivative. They are mainly synthesized in several parts of the body, including the pineal gland. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Biopterins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Biopterin
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Pyrimidine
- Vinylogous amide
- Heteroaromatic compound
- Ketimine
- Secondary alcohol
- Azacycle
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Secondary amine
- Polyol
- Amine
- Organopnictogen compound
- Primary amine
- Primary alcohol
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Imine
- Organic oxygen compound
- Organic nitrogen compound
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03dr-9220000000-602126fb02cae16969f6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0aw9-3292200000-e6b52358cd355e939ab0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_31) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_32) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_33) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_4_34) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 2V, positive | splash10-0a4i-0090000000-c40ef232ef8c10af6b4a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 3V, positive | splash10-0a4i-0090000000-40779978322477cf298a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 5V, positive | splash10-0a4i-0090000000-e49838b4e8d652d72bdf | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 7V, positive | splash10-0a4r-0390000000-b43231e2e78443a996d7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 10V, positive | splash10-014i-0920000000-5f6d06fef1bff433cb74 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 12V, positive | splash10-014i-0900000000-1f59c6362ecd3bf94b79 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 16V, positive | splash10-014i-0900000000-5299b47c47609ee94a30 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 20V, positive | splash10-014i-1900000000-f31d91ebf5d2ad43354c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 24V, positive | splash10-01ba-3900000000-4d3e2458159bb9f65ce6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 28V, positive | splash10-00xs-6900000000-86a6eac4069432a18ae5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 33V, positive | splash10-00r7-9600000000-971378f9c783f6d800ff | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 38V, positive | splash10-014l-9200000000-687b58605bb70fc727db | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Orbitrap 46V, positive | splash10-014i-9000000000-b01418dfcc77acb66b6e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-014i-0900000000-5da76eb2e54c7dcf9422 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-014i-0900000000-df54ce2cbff970a469f4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-004i-0900000000-d6756ef30d881fb25685 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-001l-0900000000-e8baecab87622e9340c6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-0w29-0900000000-4952089b8c8b79285dea | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - n/a 17V, positive | splash10-0ab9-0690000000-1f127d1244a709e9919f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0090000000-e5dad4abb411efa4feb6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01p9-1790000000-449cabdd2508eefabccc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03dj-3900000000-b2abed21008ce5fb01bd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udl-1390000000-1e1c831e7f68ac77caed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-4950000000-948e3120b1a4f548982b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9300000000-581b6b2fd47ffbefaa0c | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|