| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:07:30 UTC |
|---|
| Update Date | 2020-05-21 16:28:46 UTC |
|---|
| BMDB ID | BMDB0004058 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
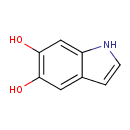
| Common Name | 5,6-Dihydroxyindole |
|---|
| Description | 5,6-Dihydroxyindole, also known as 5,6-dihydroxyindole or DHI, belongs to the class of organic compounds known as hydroxyindoles. These are organic compounds containing an indole moiety that carries a hydroxyl group. 5,6-Dihydroxyindole is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). 5,6-Dihydroxyindole participates in a number of enzymatic reactions, within cattle. In particular, 5,6-Dihydroxyindole can be biosynthesized from L-dopachrome; which is mediated by the enzyme tyrosinase. In addition, 5,6-Dihydroxyindole can be converted into indole-5,6-quinone; which is catalyzed by the enzyme tyrosinase. In cattle, 5,6-dihydroxyindole is involved in the metabolic pathway called the tyrosine metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| DHI | ChEBI | | Dopamine lutine | ChEBI | | 5,6-Dihydroxyindole | HMDB | | Indole-5,6-diol | HMDB | | Indolylquinol | HMDB |
|
|---|
| Chemical Formula | C8H7NO2 |
|---|
| Average Molecular Weight | 149.1467 |
|---|
| Monoisotopic Molecular Weight | 149.047678473 |
|---|
| IUPAC Name | 1H-indole-5,6-diol |
|---|
| Traditional Name | 5,6-dihydroxyindole |
|---|
| CAS Registry Number | 3131-52-0 |
|---|
| SMILES | OC1=C(O)C=C2C=CNC2=C1 |
|---|
| InChI Identifier | InChI=1S/C8H7NO2/c10-7-3-5-1-2-9-6(5)4-8(7)11/h1-4,9-11H |
|---|
| InChI Key | SGNZYJXNUURYCH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hydroxyindoles. These are organic compounds containing an indole moiety that carries a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Indoles and derivatives |
|---|
| Sub Class | Hydroxyindoles |
|---|
| Direct Parent | Hydroxyindoles |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydroxyindole
- Indole
- 1-hydroxy-2-unsubstituted benzenoid
- Benzenoid
- Heteroaromatic compound
- Pyrrole
- Azacycle
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-006t-0900000000-04c8c0b24d6ead789b88 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00di-4190000000-0de343d929acc2e0ab2f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-cd7bde656f825967dce0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-fc1c09de4df4722d5f82 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00sl-4900000000-88e693a5f86ce8b2fc07 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-48fdac0fa8522c16328b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-0900000000-2d3e6f8819c9c5d32c21 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kg-4900000000-c98c0942f4239d6904b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-e9c84d792f3855d1e622 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-2900000000-6476034159460897f408 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0ufu-9300000000-b351609273dd4b0ae8d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-d32a0a3b410ab4200586 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0002-1900000000-7917015f2c051992bf5a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9300000000-bade444c07fe0e0b86fe | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|