| Synonyms | | Value | Source |
|---|
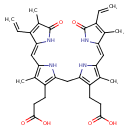
| 1,10,19,22,23,24-Hexahydro-2,7,13,17-tetramethyl-1,19-dioxo-3,18-divinylbiline-8,12-dipropionic acid | ChEBI | | 2,17-Diethenyl-1,10,19,22,23,24-hexahydro-3,7,13,18-tetramethyl-1,19-dioxo-21H-biline-8,12-dipropanoic acid | ChEBI | | 2,7,13,17-Tetramethyl-1,19-dioxo-3,18-divinyl-1,10,19,22,23,24-hexahydro-21H-biline-8,12-dipropanoic acid | ChEBI | | 8,12-Bis(2-carboxyethyl)-2,7,13,17-tetramethyl-3,18-divinylbiladiene-ac-1,19(21H,24H)-dione | ChEBI | | Bilirubin(Z,Z) | ChEBI | | Bilirubin-ixalpha | ChEBI | | 1,10,19,22,23,24-Hexahydro-2,7,13,17-tetramethyl-1,19-dioxo-3,18-divinylbiline-8,12-dipropionate | Generator | | 2,17-Diethenyl-1,10,19,22,23,24-hexahydro-3,7,13,18-tetramethyl-1,19-dioxo-21H-biline-8,12-dipropanoate | Generator | | 2,7,13,17-Tetramethyl-1,19-dioxo-3,18-divinyl-1,10,19,22,23,24-hexahydro-21H-biline-8,12-dipropanoate | Generator | | Bilirubin ixalpha | HMDB | | (4Z,15Z)-Bilirubin ixa | HMDB | | (Z,Z)-Bilirubin ixa | HMDB | | 1,10,19,22,23,24-Hexahydro-2,7,13,17-tetramethyl-1,19-dioxo-3,18-divinyl-biline-8,12-dipropionate | HMDB | | 1,10,19,22,23,24-Hexahydro-2,7,13,17-tetramethyl-1,19-dioxo-3,18-divinyl-biline-8,12-dipropionic acid | HMDB | | 3-(2-((3-(2-Carboxyethyl)-4-methyl-5-((3-methyl-5-oxo-4-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene)methyl)-1H-pyrrol-2-yl)methyl)-4-methyl-5-((4-methyl-5-oxo-3-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene)methyl)-1H-pyrrol-3-yl)propanoate | HMDB | | 3-(2-((3-(2-Carboxyethyl)-4-methyl-5-((3-methyl-5-oxo-4-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene)methyl)-1H-pyrrol-2-yl)methyl)-4-methyl-5-((4-methyl-5-oxo-3-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene)methyl)-1H-pyrrol-3-yl)propanoic acid | HMDB | | 3-(2-((3-(2-Carboxyethyl)-4-methyl-5-[(Z)-(3-methyl-5-oxo-4-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-1H-pyrrol-2-yl)methyl)-4-methyl-5-[(Z)-(4-methyl-5-oxo-3-vinyl-1,5-dihydro-2H-pyrrol-2-ylidene | HMDB | | 3-[2-[[3-(2-Carboxyethyl)-5-[(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl]methyl]-5-[(4-ethenyl-3-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoate | HMDB | | 3-[2-[[3-(2-Carboxyethyl)-5-[(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl]methyl]-5-[(4-ethenyl-3-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid | HMDB | | 3-[2-[[3-(2-Carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoate | HMDB | | 3-[2-[[3-(2-Carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid | HMDB | | Bilirubin IX-alpha | HMDB | | Cholerythrin | HMDB | | Hematoidin | HMDB | | Bilirubin IX alpha | HMDB |
|
|---|
| InChI Identifier | InChI=1S/C33H36N4O6/c1-7-20-19(6)32(42)37-27(20)14-25-18(5)23(10-12-31(40)41)29(35-25)15-28-22(9-11-30(38)39)17(4)24(34-28)13-26-16(3)21(8-2)33(43)36-26/h7-8,13-14,34-35H,1-2,9-12,15H2,3-6H3,(H,36,43)(H,37,42)(H,38,39)(H,40,41)/b26-13-,27-14- |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00vr-0100090000-d19585e898437e8be1d5 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-01vn-3100069000-c39a7113e7f8eb3fa22a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Bilirubin,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-000i-0090000000-50ff729317f07f6120b5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-0002-0090000000-83d0d13757dcbce35604 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0002-0090000000-491b2676042d62727784 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-01p9-1090000000-1d547122826037e04cfb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-000i-1090000000-30f7fa38dcf8706b38c6 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-001i-1030090000-dea3b39afc072d2855c5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-000i-1090010000-369d79a300efec3a6c78 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0091050000-78e3736d034dec9b8fa9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-0002-0091000000-cc507e875cc295800fa0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0090000000-679d7d93f8aeb5b1c0d1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0090020000-ea54f49e76225d251990 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Positive | splash10-0002-0091000000-24657f467af3adfa1e8c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-000i-0020090000-5bcce058438671178ee4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0002-0092030000-24054054aef5b235a850 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 0V, Positive | splash10-000i-0030090000-b525a9b7bb66406e6afc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00ks-0000090000-ddf9acb7f968b9079764 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00g0-0110290000-ae6f323e9f5e9e26199c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uml-1102910000-df5146b3612e4fb4ce5e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0000090000-6deb1aed1a9c3839e2a9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-053i-1010190000-7435a4388bade8e83d10 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9010240000-cc90fff0581c6bcefd0d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0010090000-d21be2c11a0dfd17c6e9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-0030190000-749f82b203e182b3e46f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0kac-0291230000-962a583eff6dd8d5cf73 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0010090000-467141ca1354a02b0aef | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, C, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|