| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:03:49 UTC |
|---|
| Update Date | 2020-06-04 20:27:49 UTC |
|---|
| BMDB ID | BMDB0003339 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | D-Glutamic acid |
|---|
| Description | D-Glutamic acid, also known as glutamate D-form or D-glutamate, belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. D-Glutamic acid exists as a solid, possibly soluble (in water), and a very strong basic compound (based on its pKa) molecule. D-Glutamic acid exists in all living organisms, ranging from bacteria to humans. |
|---|
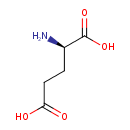
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-2-Aminopentanedioic acid | ChEBI | | D-2-Aminoglutaric acid | ChEBI | | D-Glutaminic acid | ChEBI | | D-Glutaminsaeure | ChEBI | | DGL | ChEBI | | Glutamic acid D-form | ChEBI | | (R)-2-Aminopentanedioate | Generator | | D-2-Aminoglutarate | Generator | | D-Glutaminate | Generator | | Glutamate D-form | Generator | | D-Glutamate | Generator | | (2R)-2-Aminopentanedioate | HMDB | | (2R)-2-Aminopentanedioic acid | HMDB | | D-2-Aminopentanedioate | HMDB | | D-2-Aminopentanedioic acid | HMDB | | delta-2-Aminoglutarate | HMDB | | delta-2-Aminoglutaric acid | HMDB | | delta-2-Aminopentanedioate | HMDB | | delta-2-Aminopentanedioic acid | HMDB | | delta-Glutamate | HMDB | | delta-Glutamic acid | HMDB | | delta-Glutaminate | HMDB | | delta-Glutaminic acid | HMDB | | delta-Glutaminsaeure | HMDB | | Glutamate | HMDB | | Lopac-g-2128 | HMDB | | Lopac-gamma-2128 | HMDB | | R-(-)-Glutamate | HMDB | | R-(-)-Glutamic acid | HMDB | | Tocris-0217 | HMDB | | Glutamic acid, (D)-isomer | HMDB | | L Glutamic acid | HMDB | | L-Glutamate | HMDB | | L-Glutamic acid | HMDB | | D Glutamate | HMDB | | Glutamate, potassium | HMDB | | L-Glutamate, aluminum | HMDB | | Aluminum L glutamate | HMDB | | Glutamic acid | HMDB | | L Glutamate | HMDB | | Aluminum L-glutamate | HMDB | | Potassium glutamate | HMDB |
|
|---|
| Chemical Formula | C5H9NO4 |
|---|
| Average Molecular Weight | 147.1293 |
|---|
| Monoisotopic Molecular Weight | 147.053157781 |
|---|
| IUPAC Name | (2R)-2-aminopentanedioic acid |
|---|
| Traditional Name | D-glutamic acid |
|---|
| CAS Registry Number | 6893-26-1 |
|---|
| SMILES | N[C@H](CCC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H9NO4/c6-3(5(9)10)1-2-4(7)8/h3H,1-2,6H2,(H,7,8)(H,9,10)/t3-/m1/s1 |
|---|
| InChI Key | WHUUTDBJXJRKMK-GSVOUGTGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamic acid and derivatives. Glutamic acid and derivatives are compounds containing glutamic acid or a derivative thereof resulting from reaction of glutamic acid at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamic acid and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamic acid or derivatives
- Alpha-amino acid
- D-alpha-amino acid
- Amino fatty acid
- Dicarboxylic acid or derivatives
- Fatty acid
- Fatty acyl
- Amino acid
- Carboxylic acid
- Organic oxide
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Organopnictogen compound
- Carbonyl group
- Organic oxygen compound
- Amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 201 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 8.88 mg/mL at 25 °C | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0k9x-9300000000-a43e0122a45b9abf09d6 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-00dl-9240000000-53c8c708191dfcd550d0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0900000000-ad4a2989e599a67182c0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0udi-0900000000-c449cfd2231fe941d44f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0udi-1900000000-f71458032a132069f94f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0udi-7900000000-810570c8cfcea8da64f5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-d5167570d11d77fd541e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-0900000000-c7d42b011a328bb7991d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-001i-9300000000-9fd882f84339f600b5eb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-001i-9000000000-407a30a35de9c1cceb3a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-001i-9000000000-8610ea89d52ef116caf5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0a4i-9000000000-331b7ed6ea33880c306e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-3900000000-58248bb2a0c7ddc905cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0zgi-9600000000-076b628a6858a60a475f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-bd3823a5fab8a2d9b62c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-1900000000-11233b5887638266e512 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fba-4900000000-1e8ea62e3fbd8467396f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0abc-9100000000-6c956e122b59de69a472 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f89-4900000000-c467fa409affedc9ab85 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9100000000-ddda6500978ea1670dcf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9000000000-4c6639c8639f077b57a0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0059-4900000000-503f6a47bc72662de09c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ugi-3900000000-ec06b29308ff068bf7a2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-e06c31345e8a86e71de1 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|