| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:06 UTC |
|---|
| Update Date | 2020-05-21 16:28:55 UTC |
|---|
| BMDB ID | BMDB0001178 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Adenosine diphosphate ribose |
|---|
| Description | Adenosine diphosphate ribose, also known as ADP-rib or ribose adenosine diphosphate, belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. Adenosine diphosphate ribose exists in all eukaryotes, ranging from yeast to plants to humans. Based on a literature review a small amount of articles have been published on Adenosine diphosphate ribose. |
|---|
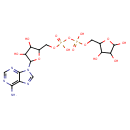
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Adenosine diphosphoric acid ribose | Generator | | (Rib5)ppa | HMDB | | 5-(Adenosine 5'-pyrophosphoryl)-D-ribose | HMDB | | a5'pp5RIb | HMDB | | Adenosine 5'-diphosphoribose | HMDB | | AdoPPRib | HMDB | | ADP Ribose | HMDB | | ADP-D-Ribose | HMDB | | ADP-Rib | HMDB | | ADP-Ribose | HMDB | | ADPribose | HMDB | | D-Ribofuranos-5-yl-ADP | HMDB | | Ribose adenosine diphosphate | HMDB | | [5-(6-Aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [hydroxy-[(3,4,5-trihydroxyoxolan-2-yl)methoxy]phosphoryl] hydrogen phosphate | HMDB | | Diphosphate ribose, adenosine | HMDB | | 5'-Diphosphoribose, adenosine | HMDB | | Adenosine diphosphoribose | HMDB | | Diphosphoribose, adenosine | HMDB | | Adenosine 5' diphosphoribose | HMDB | | Ribose, ADP | HMDB | | Ribose, adenosine diphosphate | HMDB |
|
|---|
| Chemical Formula | C15H23N5O14P2 |
|---|
| Average Molecular Weight | 559.3157 |
|---|
| Monoisotopic Molecular Weight | 559.071673493 |
|---|
| IUPAC Name | {[5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}({hydroxy[(3,4,5-trihydroxyoxolan-2-yl)methoxy]phosphoryl}oxy)phosphinic acid |
|---|
| Traditional Name | adp ribose |
|---|
| CAS Registry Number | 20762-30-5 |
|---|
| SMILES | NC1=C2N=CN(C3OC(COP(O)(=O)OP(O)(=O)OCC4OC(O)C(O)C4O)C(O)C3O)C2=NC=N1 |
|---|
| InChI Identifier | InChI=1S/C15H23N5O14P2/c16-12-7-13(18-3-17-12)20(4-19-7)14-10(23)8(21)5(32-14)1-30-35(26,27)34-36(28,29)31-2-6-9(22)11(24)15(25)33-6/h3-6,8-11,14-15,21-25H,1-2H2,(H,26,27)(H,28,29)(H2,16,17,18) |
|---|
| InChI Key | SRNWOUGRCWSEMX-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine nucleotide sugars |
|---|
| Direct Parent | Purine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine nucleotide sugar
- Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Pyrimidine
- Alkyl phosphate
- Imidolactam
- Phosphoric acid ester
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Tetrahydrofuran
- Azole
- Imidazole
- Heteroaromatic compound
- Secondary alcohol
- Hemiacetal
- Organoheterocyclic compound
- Polyol
- Azacycle
- Oxacycle
- Organic oxide
- Amine
- Alcohol
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Hydrocarbon derivative
- Organic nitrogen compound
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01r5-1196680000-fb6c20cc4fb54b50028e | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-01r5-2115904000-4597a65923641317c791 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Adenosine diphosphate ribose,1TMS,#1" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0910130000-b9073b1bbcee0d4c7c6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0910000000-4407a7719b31b18bb57b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1900000000-9d6c2b567f9da9d1d1ce | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a59-1701390000-e958d4521e35989750e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1910100000-a653cbe7cba6b5fab234 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a7i-3900000000-c8a10ccf06e1cb8b548a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0000090000-36723dbc3777092ad5f2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0faa-2853950000-c2c9d5c0b802214dc6cc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2921100000-97be795a8dc0bb3af64e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0000290000-f9ecd49245b2df56a8de | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ar0-1221970000-04d99933f777218ddd3c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a6v-5932310000-f0d589f4eccb4121906e | View in MoNA |
|---|
|
|---|