| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:31 UTC |
|---|
| Update Date | 2020-05-21 16:29:08 UTC |
|---|
| BMDB ID | BMDB0001201 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Guanosine diphosphate |
|---|
| Description | Guanosine diphosphate, also known as Guanosine diphosphate or GDP, belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. Guanosine diphosphate is possibly soluble (in water) and a moderately basic compound (based on its pKa). Guanosine diphosphate exists in all living species, ranging from bacteria to humans. In cattle, guanosine diphosphate is involved in a couple of metabolic pathways, which include the intracellular signalling through adenosine receptor A2A and adenosine pathway and the intracellular signalling through adenosine receptor A2B and adenosine pathway. |
|---|
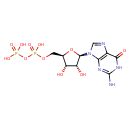
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Guanosine 5'-diphosphate | ChEBI | | Guanosine 5'-diphosphoric acid | Generator | | Guanosine diphosphoric acid | Generator | | 5'-GDP | HMDB | | GDP | HMDB | | Guanosine 5'-(trihydrogen pyrophosphate) | HMDB | | Guanosine 5'-pyrophosphate | HMDB | | Guanosine mono(trihydrogen diphosphate) | HMDB | | Guanosine pyrophosphate | HMDB | | Guanosine-5'-diphosphate | HMDB | | Guanosine-diphosphate | HMDB | | PpG | HMDB | | 5'-Diphosphate, guanosine | HMDB | | Diphosphate, guanosine 5'-trihydrogen | HMDB | | Guanosine 5' diphosphate | HMDB | | Guanosine 5' trihydrogen diphosphate | HMDB | | 5'-Trihydrogen diphosphate, guanosine | HMDB | | Diphosphate, guanosine | HMDB | | Guanosine 5'-trihydrogen diphosphate | HMDB |
|
|---|
| Chemical Formula | C10H15N5O11P2 |
|---|
| Average Molecular Weight | 443.2005 |
|---|
| Monoisotopic Molecular Weight | 443.024329371 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(2-amino-6-oxo-6,9-dihydro-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]phosphonic acid |
|---|
| Traditional Name | {[(2R,3S,4R,5R)-5-(2-amino-6-oxo-3H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxyphosphonic acid |
|---|
| CAS Registry Number | 146-91-8 |
|---|
| SMILES | NC1=NC(=O)C2=C(N1)N(C=N2)[C@@H]1O[C@H](COP(O)(=O)OP(O)(O)=O)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C10H15N5O11P2/c11-10-13-7-4(8(18)14-10)12-2-15(7)9-6(17)5(16)3(25-9)1-24-28(22,23)26-27(19,20)21/h2-3,5-6,9,16-17H,1H2,(H,22,23)(H2,19,20,21)(H3,11,13,14,18)/t3-,5-,6-,9-/m1/s1 |
|---|
| InChI Key | QGWNDRXFNXRZMB-UUOKFMHZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purine ribonucleoside diphosphates. These are purine ribobucleotides with diphosphate group linked to the ribose moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Purine nucleotides |
|---|
| Sub Class | Purine ribonucleotides |
|---|
| Direct Parent | Purine ribonucleoside diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine ribonucleoside diphosphate
- Purine ribonucleoside monophosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-oxopurine
- Hypoxanthine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Pyrimidone
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Alkyl phosphate
- Phosphoric acid ester
- Pyrimidine
- Vinylogous amide
- Tetrahydrofuran
- Azole
- Imidazole
- Heteroaromatic compound
- Secondary alcohol
- 1,2-diol
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Organooxygen compound
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Alcohol
- Amine
- Organic oxygen compound
- Primary amine
- Organonitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Cytoplasm
- Golgi
- Mitochondria
- Nucleus
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-004i-5930200000-bdc0893316c12636a495 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0229-6393250000-a57b7f18975223fa888c | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udl-0900600000-5c7173a5771dfefadc5e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0udi-0900000000-d38af29994fa5108c331 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0udi-0900000000-3072d857dcc8a1b2cbaa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-0kdi-5901100000-24a5c807e53370f01d0c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-0udi-0900000000-6399eaf97235815c4cb7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0udi-0900000000-80f8e0b42526cc03fef4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0pdl-2902100000-027fb059218f84756c35 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0pdl-2902200000-876f01fe6b2be177ac75 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0pdl-3902100000-054557b6d17ed830f7e5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9600000000-2f85040f9b280383ec8a | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-054o-5904800000-e39f64dcf91afa144498 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0001900000-cdce5e7d8986462632b2 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0kdi-5901100000-ba4bded631640c110a56 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0911300000-fcd49d8a5e9d3dc3f933 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-ded96f0f499982ac1ab0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0900000000-ac93e95f54842f42e273 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f6x-0900800000-1ed1199fdace4d1b4d80 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zir-3900000000-b9aeef613c34fe99a48c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9200000000-f630c9237e72a76d3d63 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900200000-fa2d42f6050d9ab60119 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-ed3d09503a04c850d595 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-0900000000-19a5699334c495daa4e5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0001900000-7b9b9276c5e39c703f8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9202300000-fa0fbdc8c265ee49a371 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9100000000-278ab542160fff840256 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|