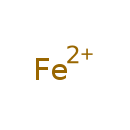Showing metabocard for Iron (BMDB0000692)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-09-30 22:34:59 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-06-04 20:47:33 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0000692 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | Iron | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | Iron, also known as fe (ii) ion or fe(ii), belongs to the class of inorganic compounds known as homogeneous transition metal compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a transition metal atom. Iron exists as a solid, possibly soluble (in water), and possibly neutral molecule. Iron exists in all living species, ranging from bacteria to humans. Iron is a potentially toxic compound. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | Fe | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 55.845 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 55.934942133 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | lambda2-iron(2+) ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | lambda2-iron(2+) ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 15438-31-0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [Fe++] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/Fe/q+2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | CWYNVVGOOAEACU-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of inorganic compounds known as homogeneous transition metal compounds. These are inorganic compounds containing only metal atoms,with the largest atom being a transition metal atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Inorganic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Homogeneous metal compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Homogeneous transition metal compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Homogeneous transition metal compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Detected and Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0015531 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB030855 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 25394 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | C14818 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | FE%2b2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | 33552 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Iron | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 27284 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | FE2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | 29033 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Only showing the first 50 proteins. There are 184 proteins in total.
Enzymes
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- MT-CO1
- Uniprot ID:
- P00396
- Molecular weight:
- 57032.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase that catalyzes the conversion of C19 androgens, androst-4-ene-3,17-dione (androstenedione) and testosterone to the C18 estrogens, estrone and estradiol, respectively. Catalyzes three successive oxidations of C19 androgens: two conventional oxidations at C19 yielding 19-hydroxy and 19-oxo/19-aldehyde derivatives, followed by a third oxidative aromatization step that involves C1-beta hydrogen abstraction combined with cleavage of the C10-C19 bond to yield a phenolic A ring and formic acid. Alternatively, the third oxidative reaction yields a 19-norsteroid and formic acid. Converts dihydrotestosterone to delta1,10-dehydro 19-nordihydrotestosterone and may play a role in homeostasis of this potent androgen. Also displays 2-hydroxylase activity toward estrone. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase).
- Gene Name:
- CYP19A1
- Uniprot ID:
- P46194
- Molecular weight:
- 58090.0
- General function:
- Involved in cytochrome-c oxidase activity
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COX5A
- Uniprot ID:
- P00426
- Molecular weight:
- 16735.0
- General function:
- Involved in dopamine beta-monooxygenase activity
- Specific function:
- Ferric-chelate reductases reduce Fe(3+) to Fe(2+) before its transport from the endosome to the cytoplasm.
- Gene Name:
- FRRS1
- Uniprot ID:
- A2VE04
- Molecular weight:
- 65660.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase involved in the biosynthesis of adrenal corticoids. Catalyzes the hydroxylation of carbon hydrogen bond at 11-beta position of 11-deoxycortisol and 11-deoxycorticosterone/21-hydroxyprogesterone yielding cortisol or corticosterone, respectively. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate and reducing the second into a water molecule. Two electrons are provided by NADPH via a two-protein mitochondrial transfer system comprising flavoprotein FDXR (adrenodoxin/ferredoxin reductase) and nonheme iron-sulfur protein FDX1 or FDX2 (adrenodoxin/ferredoxin).
- Gene Name:
- CYP11B1
- Uniprot ID:
- P15150
- Molecular weight:
- 57847.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase involved in sterol biosynthesis. Catalyzes 14-alpha demethylation of lanosterol and 24,25-dihydrolanosterol likely through sequential oxidative conversion of 14-alpha methyl group to hydroxymethyl, then to carboxylaldehyde, followed by the formation of the delta 14,15 double bond in the sterol core and concomitant release of formic acid. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase).
- Gene Name:
- CYP51A1
- Uniprot ID:
- Q4PJW3
- Molecular weight:
- 56596.0
- General function:
- Inorganic ion transport and metabolism
- Specific function:
- Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide. Promotes growth of cells.
- Gene Name:
- CAT
- Uniprot ID:
- P00432
- Molecular weight:
- 59915.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP3A28
- Uniprot ID:
- P79102
- Molecular weight:
- 58152.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex) that is part of the mitochondrial respiratory chain. The b-c1 complex mediates electron transfer from ubiquinol to cytochrome c. Contributes to the generation of a proton gradient across the mitochondrial membrane that is then used for ATP synthesis.
- Gene Name:
- MT-CYB
- Uniprot ID:
- P00157
- Molecular weight:
- 42591.0
- General function:
- Energy production and conversion
- Specific function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
- Gene Name:
- SDHC
- Uniprot ID:
- P35720
- Molecular weight:
- 18389.0
- General function:
- Inorganic ion transport and metabolism
- Specific function:
- Produces nitric oxide (NO) which is implicated in vascular smooth muscle relaxation through a cGMP-mediated signal transduction pathway. NO mediates vascular endothelial growth factor (VEGF)-induced angiogenesis in coronary vessels and promotes blood clotting through the activation of platelets.
- Gene Name:
- NOS3
- Uniprot ID:
- P29473
- Molecular weight:
- 133287.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the ubiquinol-cytochrome c oxidoreductase, a multisubunit transmembrane complex that is part of the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. The cytochrome b-c1 complex catalyzes electron transfer from ubiquinol to cytochrome c, linking this redox reaction to translocation of protons across the mitochondrial inner membrane, with protons being carried across the membrane as hydrogens on the quinol. In the process called Q cycle, 2 protons are consumed from the matrix, 4 protons are released into the intermembrane space and 2 electrons are passed to cytochrome c. Cytochrome c1 is a catalytic core subunit containing a c-type heme. It transfers electrons from the [2Fe-2S] iron-sulfur cluster of the Rieske protein to cytochrome c.
- Gene Name:
- CYC1
- Uniprot ID:
- P00125
- Molecular weight:
- 35297.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- Catalyzes the isomerization of prostaglandin H2 to prostacyclin (= prostaglandin I2).
- Gene Name:
- PTGIS
- Uniprot ID:
- Q29626
- Molecular weight:
- 56629.0
- General function:
- Involved in heme binding
- Specific function:
- Membrane-anchoring subunit of succinate dehydrogenase (SDH) that is involved in complex II of the mitochondrial electron transport chain and is responsible for transferring electrons from succinate to ubiquinone (coenzyme Q).
- Gene Name:
- SDHD
- Uniprot ID:
- Q95123
- Molecular weight:
- 17124.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMV6
- Molecular weight:
- 20164.0
- General function:
- Involved in iron ion binding
- Specific function:
- Ferric-chelate reductase that reduces Fe(3+) to Fe(2+) before its transport from the endosome to the cytoplasm. Probably uses ascorbate as electron donor (By similarity).
- Gene Name:
- CYB561A3
- Uniprot ID:
- A5D9A7
- Molecular weight:
- 29803.0
- General function:
- Signal transduction mechanisms
- Specific function:
- Mediates responses to nitric oxide (NO) by catalyzing the biosynthesis of the signaling molecule cGMP.
- Gene Name:
- GUCY1B1
- Uniprot ID:
- P16068
- Molecular weight:
- 70502.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMT7
- Molecular weight:
- 20837.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMS5
- Molecular weight:
- 21961.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMS1
- Molecular weight:
- 22058.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COI
- Uniprot ID:
- C5IS82
- Molecular weight:
- 25404.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YN12
- Molecular weight:
- 19067.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase involved in corticoid and androgen biosynthesis. Catalyzes 17-alpha hydroxylation of C21 steroids, which is common for both pathways. A second oxidative step, required only for androgen synthesis, involves an acyl-carbon cleavage. The 17-alpha hydroxy intermediates, as part of adrenal glucocorticoids biosynthesis pathway, are precursors of cortisol. Hydroxylates steroid hormones, pregnenolone and progesterone to form 17-alpha hydroxy metabolites, followed by the cleavage of the C17-C20 bond to form C19 steroids, dehydroepiandrosterone (DHEA) and androstenedione. Has 16-alpha hydroxylase activity. Catalyzes 16-alpha hydroxylation of 17-alpha hydroxy pregnenolone, followed by the cleavage of the C17-C20 bond to form 16-alpha-hydroxy DHEA. Also 16-alpha hydroxylates androgens, relevant for estriol synthesis. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase).
- Gene Name:
- CYP17A1
- Uniprot ID:
- P05185
- Molecular weight:
- 57244.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- CYP2D14
- Uniprot ID:
- Q01361
- Molecular weight:
- 56083.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase involved in the metabolism of arachidonic acid and its conjugates. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase). Acts as an omega and omega-1 hydroxylase for arachidonic acid and possibly for other long chain fatty acids. May modulate the arachidonic acid signaling pathway and play a role in other fatty acid signaling processes. May downregulate the biological activities of N-arachidonoyl-serotonin, an endocannabinoid that has anti-nociceptive effects through inhibition of fatty acid amide hydrolase FAAH, TRPV1 receptor and T-type calcium channels. Catalyzes C-2 oxidation of the indole ring of N-arachidonoyl-serotonin forming a less active product 2-oxo-N-arachidonoyl-serotonin.
- Gene Name:
- CYP2U1
- Uniprot ID:
- Q0IIF9
- Molecular weight:
- 61997.0
- General function:
- Involved in heme binding
- Specific function:
- Dual cyclooxygenase and peroxidase in the biosynthesis pathway of prostanoids, a class of C20 oxylipins mainly derived from arachidonate, with a particular role in the inflammatory response. The cyclooxygenase activity oxygenates arachidonate (AA, C20:4(n-6)) to the hydroperoxy endoperoxide prostaglandin G2 (PGG2), and the peroxidase activity reduces PGG2 to the hydroxy endoperoxide PGH2, the precursor of all 2-series prostaglandins and thromboxanes. This complex transformation is initiated by abstraction of hydrogen at carbon 13 (with S-stereochemistry), followed by insertion of molecular O2 to form the endoperoxide bridge between carbon 9 and 11 that defines prostaglandins. The insertion of a second molecule of O2 (bis-oxygenase activity) yields a hydroperoxy group in PGG2 that is then reduced to PGH2 by two electrons. Similarly catalyzes successive cyclooxygenation and peroxidation of dihomo-gamma-linoleate (DGLA, C20:3(n-6)) and eicosapentaenoate (EPA, C20:5(n-3)) to corresponding PGH1 and PGH3, the precursors of 1- and 3-series prostaglandins. In an alternative pathway of prostanoid biosynthesis, converts 2-arachidonoyl lysophopholipids to prostanoid lysophopholipids, which are then hydrolyzed by intracellular phospholipases to release free prostanoids. Metabolizes 2-arachidonoyl glycerol yielding the glyceryl ester of PGH2, a process that can contribute to pain response. Generates lipid mediators from n-3 and n-6 polyunsaturated fatty acids (PUFAs) via a lipoxygenase-type mechanism. Oxygenates PUFAs to hydroperoxy compounds and then reduces them to corresponding alcohols. Plays a role in the generation of resolution phase interaction products (resolvins) during both sterile and infectious inflammation. Metabolizes docosahexaenoate (DHA, C22:6(n-3)) to 17R-HDHA, a precursor of the D-series resolvins (RvDs). As a component of the biosynthetic pathway of E-series resolvins (RvEs), converts eicosapentaenoate (EPA, C20:5(n-3)) primarily to 18S-HEPE that is further metabolized by ALOX5 and LTA4H to generate 18S-RvE1 and 18S-RvE2. In vascular endothelial cells, converts docosapentaenoate (DPA, C22:5(n-3)) to 13R-HDPA, a precursor for 13-series resolvins (RvTs) shown to activate macrophage phagocytosis during bacterial infection. In activated leukocytes, contributes to oxygenation of hydroxyeicosatetraenoates (HETE) to diHETES (5,15-diHETE and 5,11-diHETE) (By similarity). During neuroinflammation, plays a role in neuronal secretion of specialized preresolving mediators (SPMs) 15R-lipoxin A4 that regulates phagocytic microglia (By similarity).
- Gene Name:
- PTGS2
- Uniprot ID:
- O62698
- Molecular weight:
- 69163.0
- General function:
- Involved in heme binding
- Specific function:
- Converts arachidonate to prostaglandin H2 (PGH2), a committed step in prostanoid synthesis. Involved in the constitutive production of prostanoids in particular in the stomach and platelets. In gastric epithelial cells, it is a key step in the generation of prostaglandins, such as prostaglandin E2 (PGE2), which plays an important role in cytoprotection. In platelets, it is involved in the generation of thromboxane A2 (TXA2), which promotes platelet activation and aggregation, vasoconstriction and proliferation of vascular smooth muscle cells (By similarity).
- Gene Name:
- PTGS1
- Uniprot ID:
- O62664
- Molecular weight:
- 68805.0
- General function:
- Involved in heme binding
- Specific function:
- Critical component of the membrane-bound oxidase of phagocytes that generates superoxide. Associates with NOX3 to form a functional NADPH oxidase constitutively generating superoxide.
- Gene Name:
- CYBA
- Uniprot ID:
- O46521
- Molecular weight:
- 20496.0
- General function:
- Involved in calcium ion binding
- Specific function:
- Antimicrobial agent which utilizes hydrogen peroxide and thiocyanate (SCN) to generate the antimicrobial substance hypothiocyanous acid (HOSCN). May protect the udder from infection and promote growth in newborn calves. Inhibits growth of the following bacterial species: E.coli, K.pneumoniae, P.aeruginosa, S.sonnei, S.saphrophyticus, S.epidermidis, and S.dysenteriae.
- Gene Name:
- LPO
- Uniprot ID:
- P80025
- Molecular weight:
- 80642.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COI
- Uniprot ID:
- C5IS85
- Molecular weight:
- 25438.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YN01
- Molecular weight:
- 19520.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase that catalyzes the side-chain hydroxylation and cleavage of cholesterol to pregnenolone, the precursor of most steroid hormones (PubMed:11412116). Catalyzes three sequential oxidation reactions of cholesterol, namely the hydroxylation at C22 followed with the hydroxylation at C20 to yield 20R,22R-hydroxycholesterol that is further cleaved between C20 and C22 to yield the C21-steroid pregnenolone and 4-methylpentanal (PubMed:11412116). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate and reducing the second into a water molecule. Two electrons are provided by NADPH via a two-protein mitochondrial transfer system comprising flavoprotein FDXR (adrenodoxin/ferredoxin reductase) and nonheme iron-sulfur protein FDX1 or FDX2 (adrenodoxin/ferredoxin) (PubMed:11412116).
- Gene Name:
- CYP11A1
- Uniprot ID:
- P00189
- Molecular weight:
- 60333.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- Not Available
- Gene Name:
- CYP20A1
- Uniprot ID:
- Q5E980
- Molecular weight:
- 52674.0
- General function:
- Involved in aromatase activity
- Specific function:
- Cytochromes P450 are a group of heme-thiolate monooxygenases. In liver microsomes, this enzyme is involved in an NADPH-dependent electron transport pathway. It oxidizes a variety of structurally unrelated compounds, including steroids, fatty acids, and xenobiotics.
- Gene Name:
- Not Available
- Uniprot ID:
- P22779
- Molecular weight:
- 2060.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMR7
- Molecular weight:
- 22171.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- Specifically catalyzes the 21-hydroxylation of steroids. Required for the adrenal synthesis of mineralocorticoids and glucocorticoids.
- Gene Name:
- CYP21
- Uniprot ID:
- P00191
- Molecular weight:
- 56077.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COXI
- Uniprot ID:
- A5YMW7
- Molecular weight:
- 19834.0
- General function:
- Secondary metabolites biosynthesis, transport and catabolism
- Specific function:
- A cytochrome P450 monooxygenase involved in the metabolism of fatty acids. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase). Catalyzes the hydroxylation of carbon-hydrogen bonds. Hydroxylates fatty acids specifically at the omega-1 position displaying the highest catalytic activity for saturated fatty acids. May be involved in the oxidative metabolism of xenobiotics.
- Gene Name:
- CYP2E1
- Uniprot ID:
- O18963
- Molecular weight:
- 56827.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COI
- Uniprot ID:
- A1XP62
- Molecular weight:
- 8746.0
- General function:
- Energy production and conversion
- Specific function:
- Component of the cytochrome c oxidase, the last enzyme in the mitochondrial electron transport chain which drives oxidative phosphorylation. The respiratory chain contains 3 multisubunit complexes succinate dehydrogenase (complex II, CII), ubiquinol-cytochrome c oxidoreductase (cytochrome b-c1 complex, complex III, CIII) and cytochrome c oxidase (complex IV, CIV), that cooperate to transfer electrons derived from NADH and succinate to molecular oxygen, creating an electrochemical gradient over the inner membrane that drives transmembrane transport and the ATP synthase. Cytochrome c oxidase is the component of the respiratory chain that catalyzes the reduction of oxygen to water. Electrons originating from reduced cytochrome c in the intermembrane space (IMS) are transferred via the dinuclear copper A center (CU(A)) of subunit 2 and heme A of subunit 1 to the active site in subunit 1, a binuclear center (BNC) formed by heme A3 and copper B (CU(B)). The BNC reduces molecular oxygen to 2 water molecules using 4 electrons from cytochrome c in the IMS and 4 protons from the mitochondrial matrix.
- Gene Name:
- COX1
- Uniprot ID:
- B1NZU1
- Molecular weight:
- 57018.0
- General function:
- Coenzyme transport and metabolism
- Specific function:
- NADH-cytochrome b5 reductase involved in endoplasmic reticulum stress response pathway. Plays a critical role in protecting pancreatic beta-cells against oxidant stress, possibly by protecting the cell from excess buildup of reactive oxygen species (ROS) (By similarity).
- Gene Name:
- CYB5R4
- Uniprot ID:
- Q32LH7
- Molecular weight:
- 59274.0
- General function:
- Involved in holocytochrome-c synthase activity
- Specific function:
- Links covalently the heme group to the apoprotein of cytochrome c.
- Gene Name:
- HCCS
- Uniprot ID:
- A5PJG7
- Molecular weight:
- 31069.0
- General function:
- Inorganic ion transport and metabolism
- Specific function:
- Produces nitric oxide (NO) which is a messenger molecule with diverse functions throughout the body. In macrophages, NO mediates tumoricidal and bactericidal actions. Also has nitrosylase activity and mediates cysteine S-nitrosylation of cytoplasmic target proteins such PTGS2/COX2. As component of the iNOS-S100A8/9 transnitrosylase complex involved in the selective inflammatory stimulus-dependent S-nitrosylation of GAPDH implicated in regulation of the GAIT complex activity and probably multiple targets including ANXA5, EZR, MSN and VIM. Involved in inflammation, enhances the synthesis of proinflammatory mediators such as IL6 and IL8.
- Gene Name:
- NOS2
- Uniprot ID:
- Q27995
- Molecular weight:
- 131208.0
- General function:
- Involved in heme oxygenase (decyclizing) activity
- Specific function:
- Heme oxygenase cleaves the heme ring at the alpha methene bridge to form biliverdin. Biliverdin is subsequently converted to bilirubin by biliverdin reductase. Under physiological conditions, the activity of heme oxygenase is highest in the spleen, where senescent erythrocytes are sequestrated and destroyed. Exhibits cytoprotective effects since excess of free heme sensitizes cells to undergo apoptosis.
- Gene Name:
- HMOX1
- Uniprot ID:
- Q5E9F2
- Molecular weight:
- 32940.0
- General function:
- Energy production and conversion
- Specific function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
- Gene Name:
- NDUFS7
- Uniprot ID:
- P42026
- Molecular weight:
- 23771.0
- General function:
- Energy production and conversion
- Specific function:
- Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I) that is believed to belong to the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
- Gene Name:
- NDUFS2
- Uniprot ID:
- P17694
- Molecular weight:
- 52556.0
- General function:
- Involved in iron ion binding
- Specific function:
- Not Available
- Gene Name:
- Not Available
- Uniprot ID:
- A9ZTK8
- Molecular weight:
- 14355.0
- General function:
- Signal transduction mechanisms
- Specific function:
- Not Available
- Gene Name:
- PPP3CC
- Uniprot ID:
- A5D7T5
- Molecular weight:
- 58180.0
- General function:
- Involved in iron ion binding
- Specific function:
- Not Available
- Gene Name:
- PPP3CB
- Uniprot ID:
- A6H703
- Molecular weight:
- 59097.0
Only showing the first 50 proteins. There are 184 proteins in total.